voluba-mriwarp – Aligning human MRI volumes to atlas space
A desktop application for aligning human MRI scans to MNI152 template and analyzing brain regions.
- Anchor your MRI scan to MNI152 template without uploading it to an online service.
- Automatically apply a set of predefined or user-defined parameters to perform nonlinear registration.
- Interactively analyze points in your MRI scan through probabilistic assignments of coordinates to brain regions of the Human Brain Atlas.
- Export anatomical assignments together with linked multimodal data features to a PDF report.
voluba-mriwarp enables you to integrate human whole-brain MRI scans into the detailed anatomical context of the Human Brain Atlas on your local computer. The tool automatically performs registration based on predefined parameters. The results can be used to assign anatomical locations to brain regions of the atlas. To perform a more detailed analysis, you can export assignments to a PDF report together with linked features like receptor densities or brain connectivity.
Integrating human MRI volumes into the Human Brain Atlas
Warping neuroscience data to a standardized space such as the ICBM MNI152 2009c nonlinear asymmetric template enables spatial integration to the Human Brain Atlas. However, the proper configuration and use of tools for skull stripping and nonlinear registration often requires significant expertise. voluba-mriwarp is a desktop application that aims to simplify both nonlinear anchoring of MRI volumes to MNI reference space, as well as interaction with the atlas, in order to assign brain regions and browse related data.
Skull-stripping and registration
To perform a detailed analysis of an MRI volume in the atlas context the input scan needs to be warped to MNI152 space. In voluba-mriwarp you can choose between different parameters for registration. If you are familiar with ANTs, you can also define your own registration parameters.
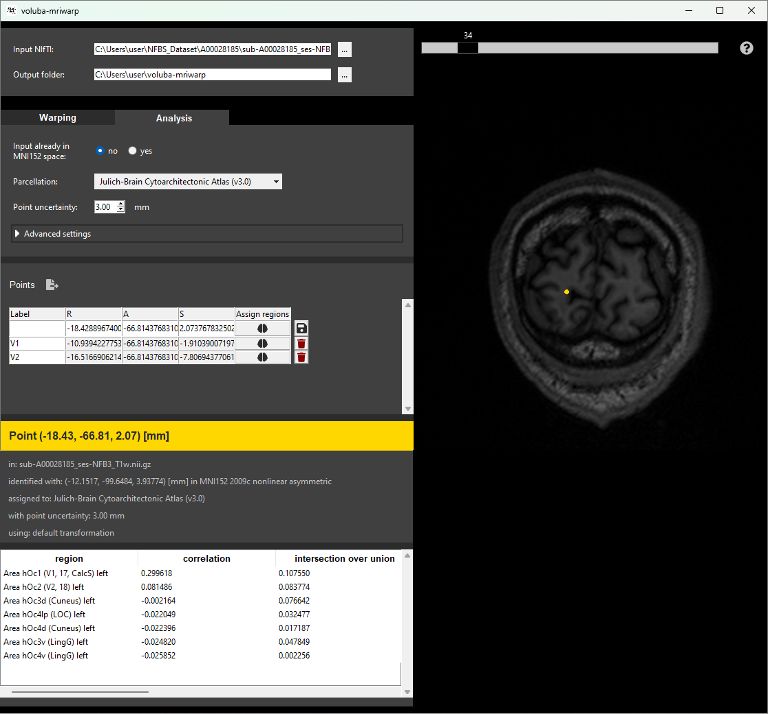
Anatomical assignment
Based on the resulting nonlinear transformation, locations in the MRI scan can be analyzed in the anatomical context of the Human Brain Atlas. You can opt for different atlas parcellations and specify a point uncertainty. Locations can be selected interactively in a viewer or entered manually into a table. voluba-mriwarp presents different measures for the selected coordinate such as a correlation coefficient or the probability values of regions in the chosen parcellation.
Extended atlas-based analysis
Each region of the assignment result can be viewed in the atlas viewer siibra-explorer. For retaining and extending the analysis, points can be saved for PDF export. The export functionality will save the anatomical assignments together with linked data features (e.g. cell density, receptor density or connectivity) for each assigned region to a PDF report.
Data safety
voluba-mriwarp is implemented as a desktop application. Therefore, MRI data will not be uploaded to any online services but remain on your local computer.
voluba-mriwarp
InstallOnline documentation
Read hereStep-by-step example
Go to the tutorial