
Complementary approaches to mapping the human brain
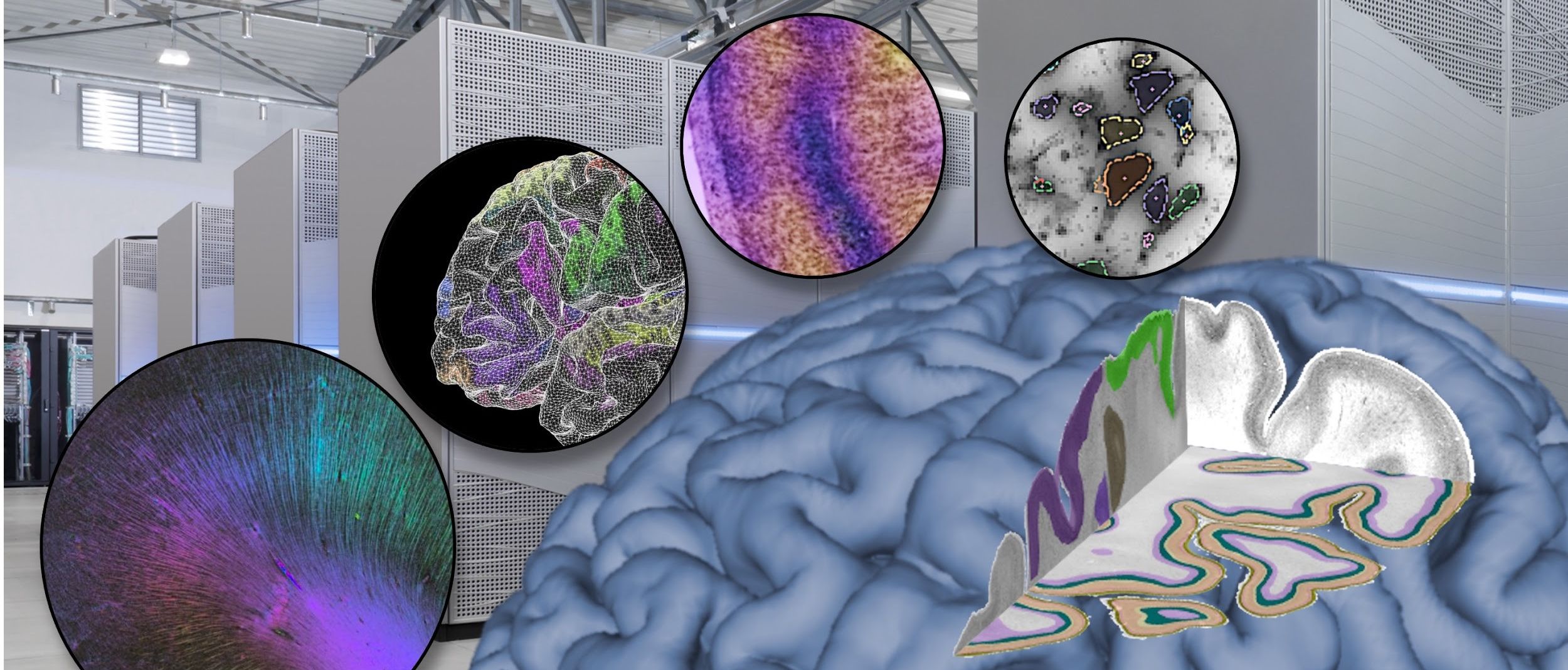
There are different approaches to brain mapping, where each reflects different modalities in brain organisation. The Human Brain Project (HBP), which concluded in September, has developed a multilevel Human Brain Atlas that offers information on cellular and molecular architecture, function, and connectivity across multiple modalities and spatial scales, i.e., is multi-level and multimodal. The Atlas is freely available on the EBRAINS platform, offering users functionalities to work with the maps, download them, and connect them to other tools, such as simulation. In addition, scientists from the HBP have also contributed to other international research efforts related to brain mapping – for example, within the US National Institutes of Health’s BRAIN initiative Cell Census Network (BICNN), which is mapping the different cell types that make up the human brain. Other mapping results include a proteomic atlas of different brain regions published this month, which profiles proteins that are involved in many cellular processes and are the target of neurological drugs.
But why do we need so many atlases and what are the differences between them?
The EBRAINS Multilevel Human Brain Atlas provides microstructurally defined anatomical areas as an interface to integrate other data types within the same reference space. The atlas, produced by HBP researchers in Germany, offers more than 250 3D maps of anatomical areas - more than ever mapped before – based on their differences in cytoarchitecture, meaning the distribution, density and morphology of cells in a three-dimensional space. It includes probability maps, which capture how variable these areas are, accounting for differences between individuals. It also includes the BigBrain model, which serves as the only available microstructural reference brain and template space.
Additionally, functional data have been integrated into the atlas, characterising brain regions based on their role in specific cognitive function.
Connectivity data reveals the intricacy of fibre tracts at millimetre resolution, and was generated through a collaboration between HBP researchers in France and Chile, based on diffusion MRI.
The atlas also features fine-scale connectivity maps at the micrometre resolution, obtained by Polarized Light imaging (PLI), and maps of the receptor distributions in the tissue mapped with quantitative Receptor Autoradiography.
Besides the human brain atlas, EBRAINS also offers detailed atlases for the macaque monkey, rat, and mouse brain in the same atlas framework.
Complementarity is key
In contrast, the US NIH initiative uses transcriptomic classification, focusing on genetic and cell type information. Their mapping is akin to a “cell census”, characterising cell types with high detail for sample sites in selected regions, but not yet with the spatial context that 3D full-brain coverage in microscopic detail would provide.
To generate a more and more complete picture of the complexity of the human brain, it becomes increasingly important to link data from different resources worldwide, and to make them interoperable. For example, EBRAINS has a tool called JuGex that allows researchers to integrate genetic information coming from the Allen brain atlas into the EBRAINS atlas, projecting the data into a common reference space, thus, promoting complementarity between the two initiatives.
“Brain atlases presenting multiple modalities, crossing the different scales, are coming of age,” comments HBP director Katrin Amunts. “Large-scale collaboration and open data sharing bring important progress. We are happy that Europe has made a significant contribution to this international neuroscience effort both with the EBRAINS Human Brain Atlas as well as with collaborations with the US.”
Cell Census
Scientists from the HBP recently collaborated with international consortia, brought together by the US National Institutes of Health’s BRAIN Initiative Cell Census Network (BICCN) to map cell types in the human brain. The studies were featured in special issues of Science, Science Advances and Science Translational Medicine in October. HBP scientists contributed to five of the 21 studies published across the journals. More than 3,000 human brain cell types were characterised through this collaborative research. This resource can help neuroscientists to model human brain diseases using other species in a more precise way.
A team at the Vrije Universiteit Amsterdam, led by HBP researcher Huib Mansvelder, contributed to four studies of the special issues. The researchers studied the consequences of gene expression for cell shape and function.
A team at University of Florence led by HBP scientist Francesco Pavone contributed to a high-resolution cellular census of Broca’s area in the brain, a region known for its important role for language. The result was published in Science Advances.
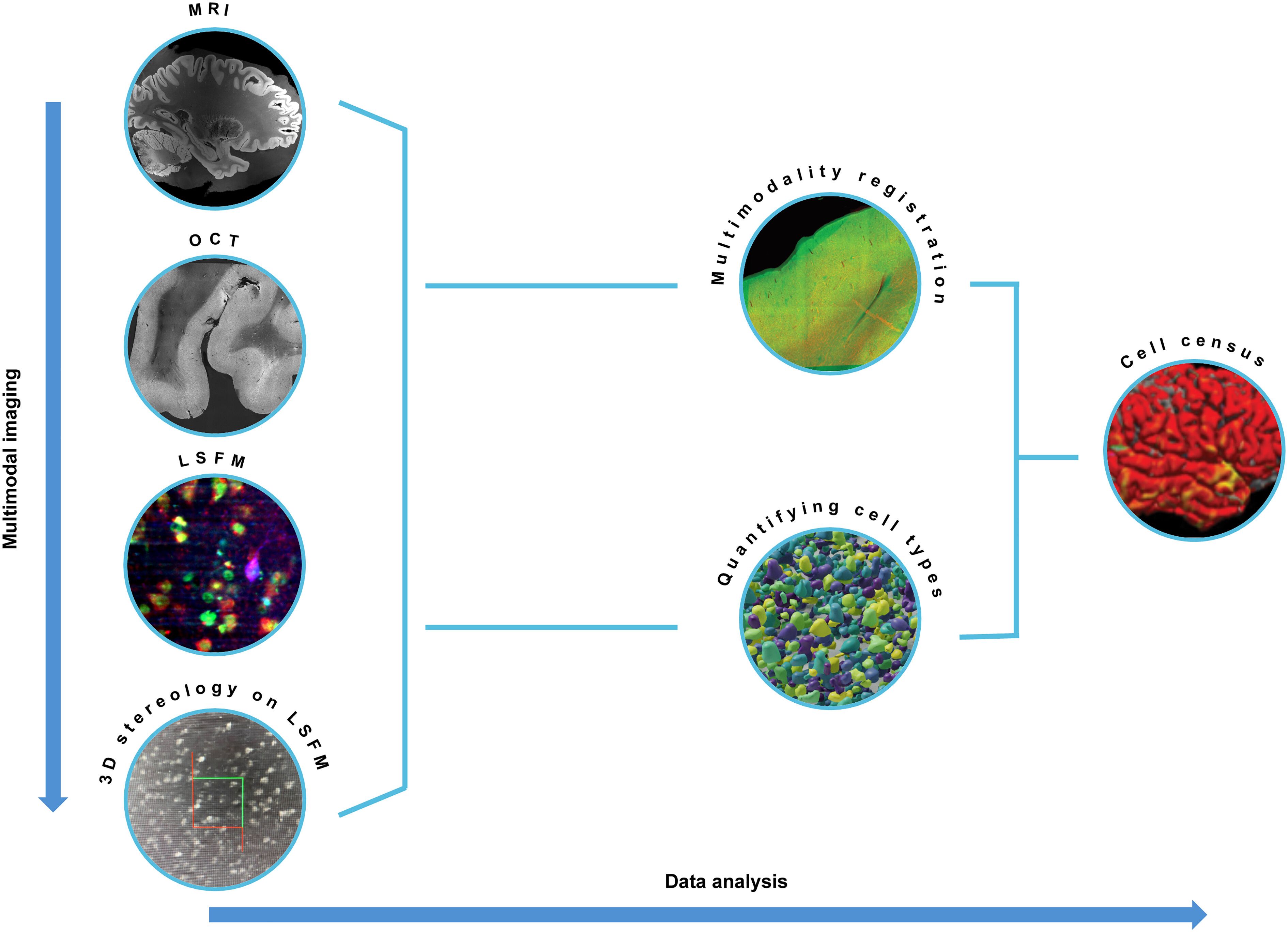
Another study published in the special issues of Science illustrates the practical applications of digital research tools offered by EBRAINS. Researchers at the Allen Institute for Brain Science, studying the organisation of the human neocortex, used the Julich Brain Maximum Probability Maps of the EBRAINS Multilevel Human Brain Atlas to map cortical tissue samples. The structures were then cross-referenced to the publicly available Julich-Brain map in the same 3D reference volume, using the Siibra-Explorer, an EBRAINS tool for atlases visualisation and navigation.
Based on FAIR data and interoperability between the different atlas approaches, along with their complementarity, they will collectively play an increasing role in pushing neuroscience forward and enabling a more precise understanding of the brain.
Read the articles with contributions from the HBP on Science:
Morphoelectric and transcriptomic divergence of the layer 1 interneuron repertoire in human versus mouse neocortex
Chartrand et al., Science 382, eadf0805 (2023) 13 October 2023
www.science.org/doi/10.1126/science.adf0805
Signature morpho-electric properties of diverse GABAergic interneurons in the human neocortex
Lee et al., Science 382, eadf6484 (2023) 13 October 2023
www.science.org/doi/10.1126/science.adf6484
Read the articles with contributions from the HBP on Science Advances:
A cellular resolution atlas of the Broca’s Area
Costantini et al., Sci. Adv. 9, eadg3844 (2023) 12 October 2023
www.science.org/doi/10.1126/sciadv.adg3844
Human voltage-gated Na+ and K+ channel properties underlie sustained fast AP signaling in human cortical neurons
Wilbers et al., Sci. Adv. 9, eade3300 (2023) 12 October 2023
www.science.org/doi/10.1126/sciadv.ade3300
Structural and functional specializations of human fast spiking neurons support fast cortical signaling
Wilbers et al., Sci. Adv. 9, eadf0708 (2023) 12 October 2023
www.science.org/doi/10.1126/sciadv.adf0708


