Subcellular Simulation Webapp
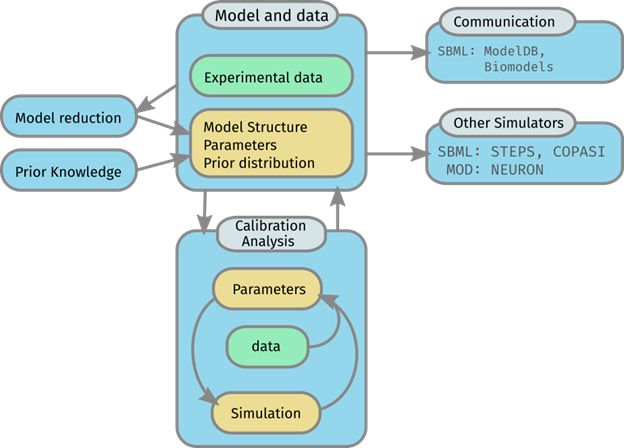
This tool allows import of SBML model files from the subcellular model building and calibration toolset workflow or other external sources. The tool allows users to setup and configure BioNetGen and STEPS simulations. Users can populate mesh models of spines and other neural structures, and run stochastic simulations of signalling pathways.