
Arbor neural network simulation library gets an update
Arbor is a multi-compartment neural network simulation library designed to be portable across contemporary high-performance computing architectures. The most recent version is now available on EBRAINS and offers the following:
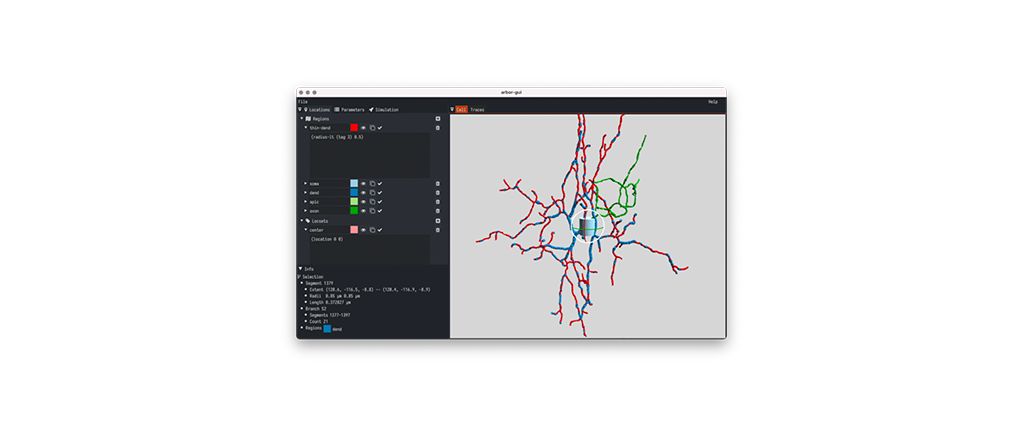
Arbor-GUI
A comprehensive tool for visually and interactively building single cell models using Arbor. It is self-contained, fast, and easy to use, and lets you peek into Arbor's brain while it is simulating the configuration you have set up. It lets the user define regions and locsets on their cells, place and parametrize ion dynamics, and manipulate cable cell parameters.
Furthermore, the simulation itself can be manipulated: users can change the timestep and runtime, add probes, stimuli and detectors, perform discretization, and finally observe traces on probes as the simulation runs. A new tutorial is included.
Mechanism ABI
This new feature provides a common C linkage ABI for externally compiled mechanisms, ion dynamics, for both CPU and GPU. The ABI allows users to create mechanisms in any language they choose, and compile them separately from Arbor and include them wherever they execute Arbor simulations. In addition, this simplifies the maintenance of mechanism "catalogues", so a user can easily package, maintain, disseminate and archive mechanisms; within their lab or with the wider world. The Brain and Behavioral Sciences Laboratory in Pavia, Italy is using this feature to interoperate with their existing mechanism collection.
User-defined gap junction models
Users can now write custom mechanisms for gap junctions, improving the ability to model synaptic plasticity.
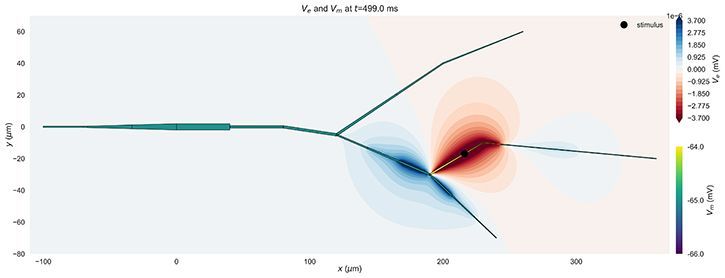
LFPykit integration
Arbor can now be used as a backend for local field potential calculations. Helper classes ArborCellGeometry and ArborLineSourcePotential are offered as part of a tutorial showing off the new feature.
Python Package
The Arbor Python Package now comes included with SWC .swc, NeuroML .nml, NeuroLucida .asc morphology support, enabling users to work with the vast majority of morphology data out there.
News & events
All news & events- Science and technology20 Dec 2024


- News20 Dec 2024

