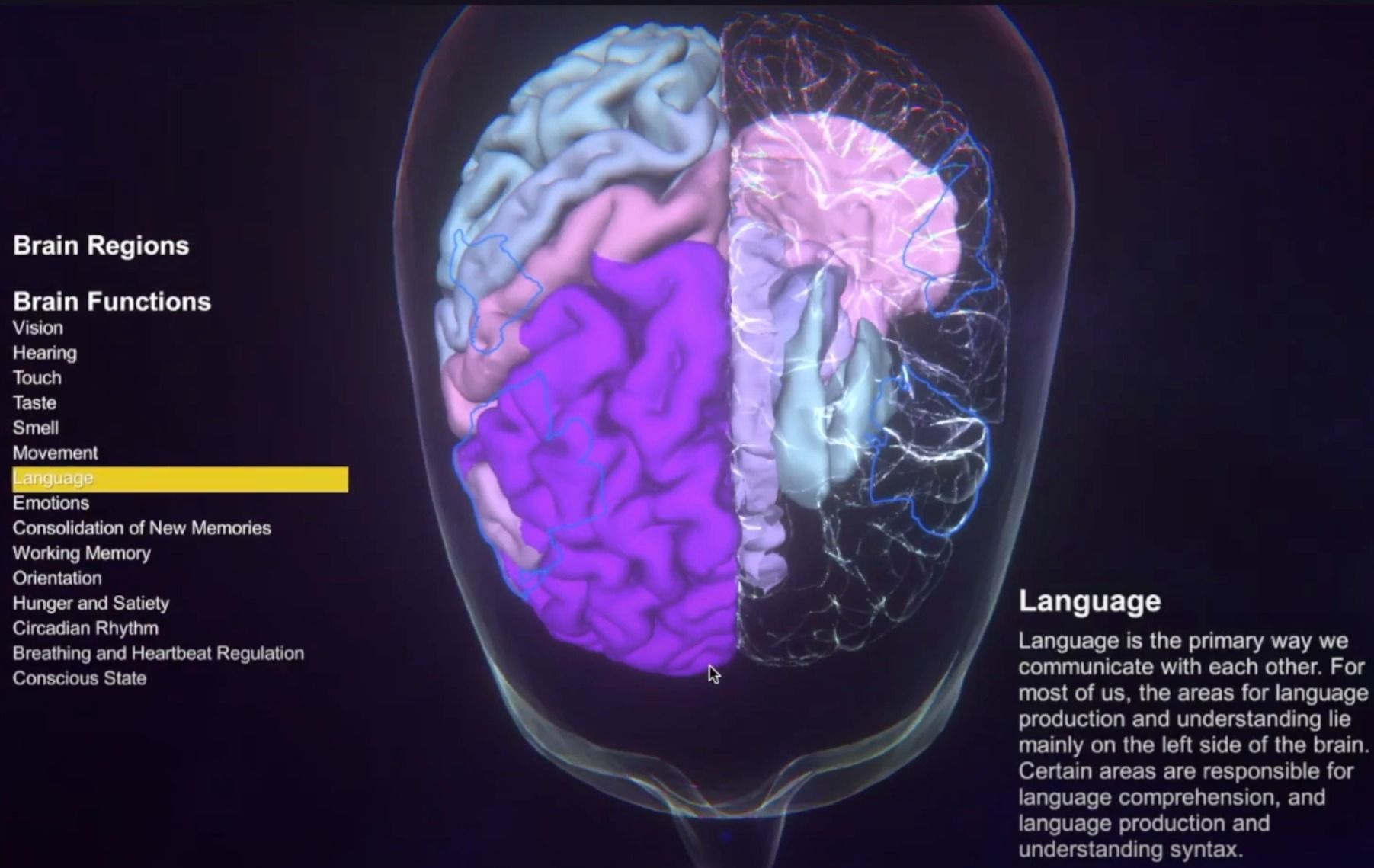
TVB Brain Atlas Viewer
A viewer that allows users to view brain atlasses on top of a 3d brain model. The Brain Atlas Viewer allows users to inspect the location and shape of different brain regions and their associated function. Brain regions can be selected by anatomy or by function. Descriptions are available in English, Arabic, Hebrew, and German. Regions are annotated with their function. TVB Brain Atlas Viewer is an interactive software that can be operated via touch screen. It was part of the HBP Travelling Exhibition that started in July 2019 at Bloomfield Museum in Jerusalem organized by the HBP Museum Program (SP11). An overview over TVB-on-EBRAINS services is provided in the preprint https://arxiv.org/abs/2102.05888