2D section images to 3D atlas
Tools for anatomical landmark based registration of mouse and rat 2D brain section images to 3D reference atlases.
Registration tools for 2D histological rodent brain section images to a 3D reference atlas
- Efficient semiautomatic affine registration of serial 2D images to a 3D reference atlas.
- Optimise initial registration using non-linear adjustments.
- Easily customise atlas overlay plates for your serial sections.
- Save the 3D spatial locations of all your sections in the atlas space for further spatial analyses.
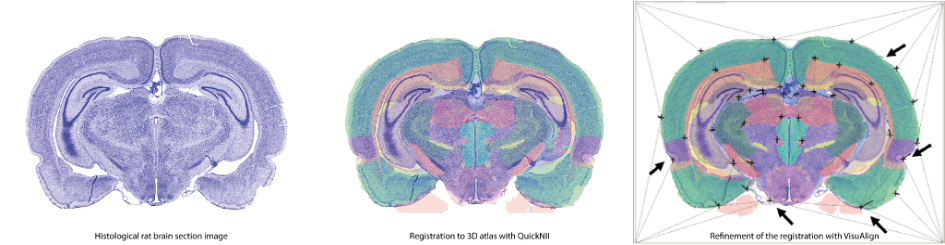
Registering different experimental neuroscience datasets to the same reference atlas allows you to spatially integrate, analyse and navigate these datasets within a standardised coordinate system. 2D histological section images of rat or mouse brains can be quickly and easily spatially registered to 3D reference atlases using QuickNII; furthermore, this registration can be fine-tuned using non-linear adjustments in VisuAlign. Both tools are downloadable desktop applications with user-friendly graphical interface and tutorials for getting started. Online versions of these tools, WebAlign and WebWarp are now also available on the EBRAINS platform.
Register mouse and rat 2D brain images to a 3D reference atlas
You can use QuickNII and VisuAlign for:
- Identification of brain regions in histological brain section image series
- Mapping the position of 3D reconstructed neurons
- Cell count analysis using the QUINT workflow
Participate in the 2D image registration community
Are you interested in atlas based spatial quantification? Discover our registration tools and interact with us for personalised advice on how to integrate your neuroscience data with atlases from mice and rats.

Post us questions directly through github.
Github QuickNII
Github VisuAlign
Course Neuroscience data integration
Related publications
Create an account
EBRAINS is open and free. Sign up now for complete access to our tools and services.