BioExcel Building Blocks
A software library for interoperable biomolecular simulation workflows.
- Collection of biomolecular workflows to explore the flexibility and dynamics of CNS macromolecules.
- Jupyter Notebooks available in the EBRAINS infrastructure, including all dependencies (ready to be run, no need of dependency tool or library installation).
- 3D graphical and interactive representation of intermediate results (NGLview, Plotly).
BioExcel Building Blocks Workflows is a collection of biomolecular workflows to explore the flexibility and dynamics of macromolecules, including signal transduction proteins or molecules related to the Central Nervous System. Molecular dynamics setup for protein and protein-ligand complexes are examples of workflows available as Jupyter Notebooks. The workflows are built using the BioBB software library, developed in the framework of the BioExcel Centre of Excellence. BioBBis a collection of Python wrappers on top of popular biomolecular simulation tools, offering a layer of interoperability between the wrapped tools, which make them compatible and prepared to be directly interconnected to build complex biomolecular workflows.
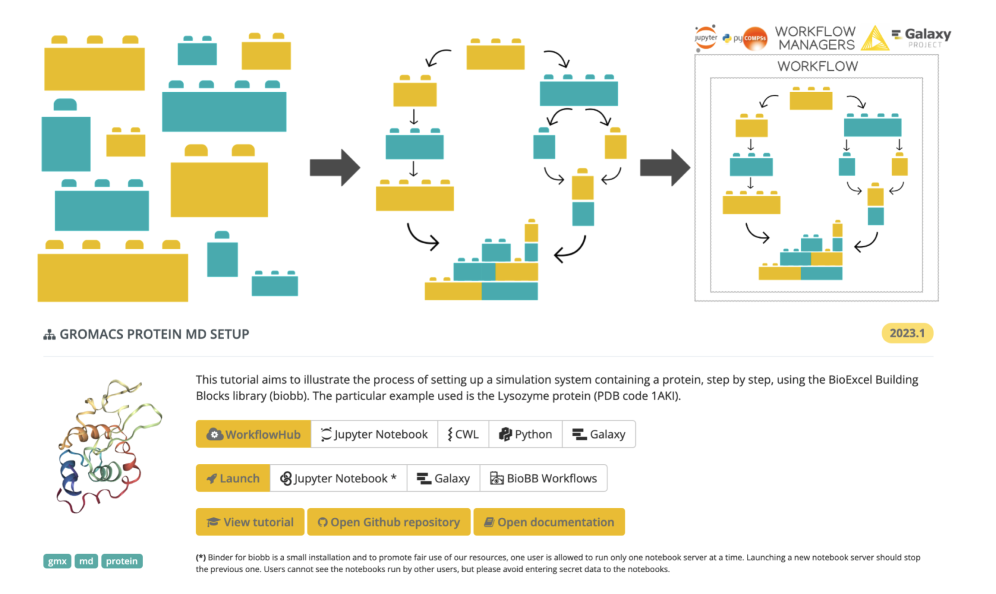
BioExcel Building Blocks (BioBB) library giving interoperability to biomolecular simulation tools, and associated workflows (e.g. Protein MD setup with GROMACS MD engine).
Other Tools
See all toolsCreate an account
EBRAINS is open and free. Sign up now for complete access to our tools and services.