MoDEL_CNS
Web-access to atomistic molecular dynamics trajectories for relevant signal transduction proteins.
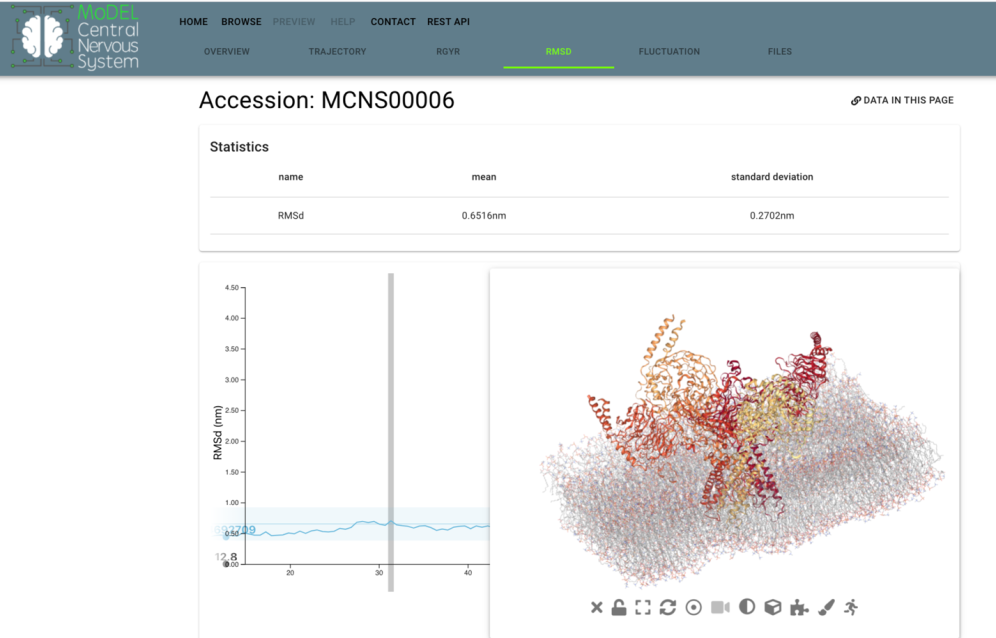
MoDEL_CNS is a platform designed to provide web-access to atomistic molecular dynamics trajectories for relevant signal transduction proteins. MoDEL_CNS expands the Molecular Dynamics Extended Library (MoDEL) database of atomistic Molecular Dynamics trajectories with proteins involved in Central Nervous System (CNS) processes, including membrane proteins. MoDEL_CNS web server interface presents the resulting trajectories, analyses, and protein properties. All data produced by the project is available to download. MoDEL_CNS will contribute to the improvement of the understanding of neuronal signalling cascades by protein structure-based simulations, calculating molecular flexibility and dynamics, and guiding systems level modelling.
Other Tools
See all toolsCreate an account
EBRAINS is open and free. Sign up now for complete access to our tools and services.