Subcellular model building and calibration tool set
Toolset for data-driven building of subcellular biochemical signaling pathway models.
The toolset includes interoperable modules for: model building, calibration (parameter estimation) and model analysis. All information needed to perform these tasks are stored in a structured, human- and machine-readable file format based on SBtab. This information includes: models, experimental calibration data and prior assumptions on parameter distributions. The toolset enables simulations of the same model in simulators with different characteristics, e.g. STEPS, NEURON, MATLAB’s Simbiology and R via automatic code generation.
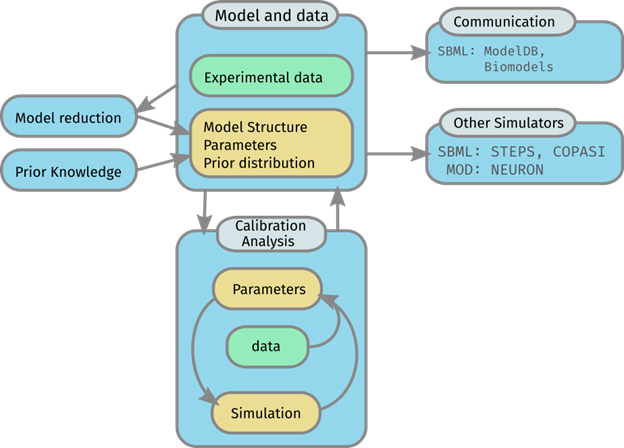
The parameter estimation is done by optimization or Bayesian approaches. Model analysis includes global sensitivity analysis and functionality for analyzing thermodynamic constraints and conserved moieties.
Documentation and source code
Uncertainty quantification and Sensitivity analysis examples
EBRAINS Jupyter Notebook example
Other Tools
See all toolsCreate an account
EBRAINS is open and free. Sign up now for complete access to our tools and services.