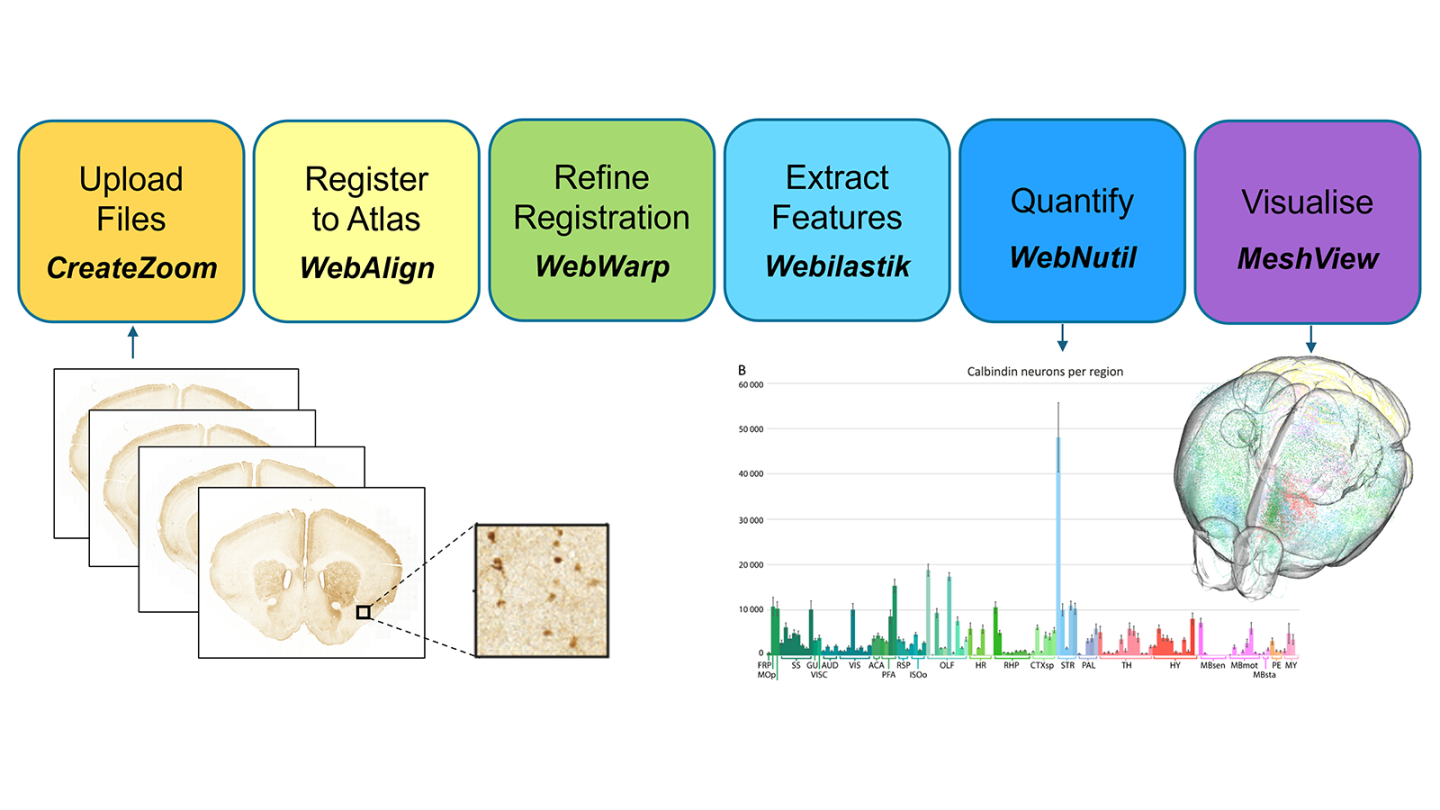
The QUINT online workflow is an analytical solution for rodent brain histology data, combining the use of a series of web-applications integrated in an online platform, for performing registration to a reference atlas, extracting features to quantify and for brain-wide mapping and quantification.
Histological studies in wild type animals or rodent disease models are used to characterise neuroanatomy, neural connections and brain diseases. Such studies benefit from section image registration to a reference brain atlas, allowing comparison and integration of disparate data within a standardised coordinate system. While analytical tools using reference brain atlases are available to researchers for histological brain section analysis, there are barriers to uptake, such as coding skill requirements, data type limitations, and insufficient documentation. The QUINT online workflow was developed to overcome such barriers, allowing researchers to register heterogeneous image series to reference brain atlases, to identify features to quantify, and to perform brain-wide mapping and quantification, in a user-friendly platform. The QUINT online workflow combines the use of a series of integrated web-applications, accessible with an EBRAINS account (CreateZoom, WebAlign, WebWarp, Webilastik, WebNutil and MeshView). It builds on the QUINT workflow (Yates et al. 2019, doi: 10.3389/fninf.2019.00075) using desktop applications, which has been used widely by the research community for cell counting, tract tracing studies and for analysis of tissue clearing data. The QUINT online workflow benefits from simplified file management steps and new features such as shareable microscopy viewer links and automated plotting. It facilitates transparent sharing of research findings through the EBRAINS data sharing platform. EBRAINS users can already access a beta version here.
Create an account
EBRAINS is open and free. Sign up now for complete access to our tools and services.