Extract, quantify and analyse features from rodent histological images
The QUINT workflow takes you through steps to quantify and analyse labelled features within a known atlas space.
- Leverage a streamlined, tested workflow for analysing labelled features in large numbers of histological sections from mouse or rat
- Obtain quantitative measures in atlas-defined regions
- Customise the granularity of regions-of-interest to fit the needs of your analysis
The QUINT workflow comprises a suite of software designed to support atlas-based quantification. All the software have user interfaces, with no coding ability required. It generates object counts and percentage coverage per atlas-region, in addition to point clouds for visualising the features in 3D.
It currently supports quantification relative to the following atlases:
- Allen Mouse Brain Atlas Common Coordinate Framework version 3 (2015 and 2017) (CCFv3)
- Waxholm Atlas of the Sprague Dawley rat, version 2, 3 and 4 (WHS rat brain atlas).
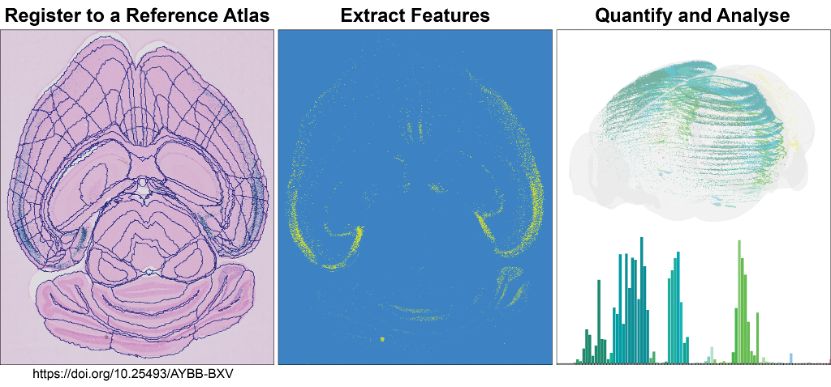
Analyse large amounts of histological image data from rat or mouse brains
Nutil
Learn moreilastik
Learn moreQuickNII
Learn moreVisuAlign
Learn moreMeshView
Learn moreQCAlign
Learn moreUse QUINT for:
- Cell count analysis
- Analysis of tract-tracing connections
- Resizing, rotating and renaming large Tiff images
Get involved in the QUINT community
Are you interested in atlas based spatial quantification? Discover the QUINT workflow and interact with us to customise the workflow for your needs: Count cells, AD plaques and quantify connectivity traces in 2D images from mice and rat. The QUINT workflow is highly flexible and has developed over time in response to users’ needs. We look forward to continuing this collaboration with the research community and welcome all requests, big and small. Post us questions directly through github or at https://ebrains.eu/support/
Try the new pre-released online QUINT workflow
We have developed an online version of the QUINT workflow for histological rodent brain image analysis. The tools are presented in a workbench were users can work in their private space. Try the beta version here: https://rodent-brain-workbench.apps.hbp.eu/
This is a pre-release, if you get into issues, contact us at support@ebrains.eu.
References
- Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., et al. (2019) ilastik:interactive machine learning for (bio) image analysis. Nat Methods. 16, 1226–1232. https://doi.org/10.1038/s41592-019-0582-9
- Groeneboom N.E., Yates S.C., Puchades M.A. and Bjaalie J.G. (2020) Nutil: A Pre- and Post-processing Toolbox for Histological Rodent Brain Section Images. Front. Neuroinform. 14:37. https://doi.org/10.3389/fninf.2020.00037
- Puchades M.A., Csucs G., Lederberger D., Leergaard T.B., and Bjaalie J.G. (2019) Spatial registration of serial microscopic brain images to three-dimensional reference atlases with the QuickNII tool. PLosONE. 14 (5): e0216796. https://doi.org/10.1371/journal.pone.0216796
- Yates S.C., Groeneboom N.E., Coello C., Lichtenthaler S.F., Kuhn P.H., Demuth H.U., Hartlage-Rübsamen M., Roßner S., Leergaard T., Kreshuk A., Puchades M.A., Bjaalie JG. (2019) QUINT: Workflow for quantification and spatial analysis of features in histological images from rodent brain. Front. Neuroinform. 13, 75. https://doi.org/10.3389/fninf.2019.00075